This post is in a series on machine learning with unbalanced datasets. This post focuses on the makeup of the validation set in particular. For background, please see the setup post.
Table of Contents
- Train
- Experiment #1
- Experiment #2
- Experiment #3
- Round 1 Conclusion
- Experiment #4
- Experiment #5
- Experiment #6
- Overall Conclusion
%run 2021-05-01-experiments-on-unbalanced-datasets-setup.ipynb
Classes:
['cats', 'dogs']
There are a total of 5000 cat images in the entire dataset.
There are a total of 5000 dog images in the entire dataset.
Train
Let’s start out by creating our datasets. We’ll make a balanced train set and a representative test set and keep those constant. We’ll also make two validation sets - one will be balanced between the classes and the other will be representative of the “real world”.
It might seem weird to do this with the validation sets - shouldn’t it be the same distribution as the test set and thus the real world? Yes, absolutely it should, but we have to keep in mind that we’re often doing double work with our validation sets. We’ll use them to validate our results, but sometimes we also use them to guide our training through callbacks. For example, if we use ReduceLROnPlateau or EarlyStopping, we have to be aware that if the validation set isn’t balanced, the loss might not always decrease as expected.
This makes me want to see what balance of validation sets produces better end results on the test set.
cat_list_train, cat_list_val_balanced, cat_list_val_representative, cat_list_test = subset_dataset(
cat_list_ds, [2000, 1000, 1000, 1000]
)
dog_list_train, dog_list_val_balanced, dog_list_val_representative, dog_list_test = subset_dataset(
dog_list_ds, [2000, 1000, 100, 100]
)
train_ds = prepare_dataset(cat_list_train, dog_list_train)
val_ds_balanced = prepare_dataset(cat_list_val_balanced, dog_list_val_balanced)
val_ds_representative = prepare_dataset(cat_list_val_representative, dog_list_val_representative)
test_ds = prepare_dataset(cat_list_test, dog_list_test)
Now let’s set up the callbacks.
callbacks = [
tf.keras.callbacks.ReduceLROnPlateau(patience=2)
]
I’ll do three different experiments. They’ll be identical except for the learning rate. I want to vary the learning rate because this greatly affects model convergence, and I want to make sure the results we get are robust.
Experiment #1
optimizer = tf.keras.optimizers.Adam(learning_rate=0.0003)
history_balanced, preds_balanced, evals_balanced = run_experiment(
train_ds, val_ds_balanced, test_ds, callbacks=callbacks
)
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 3s 39ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 3s 37ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
history_representative, preds_representative, evals_representative = run_experiment(
train_ds, val_ds_representative, test_ds, callbacks=callbacks
)
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
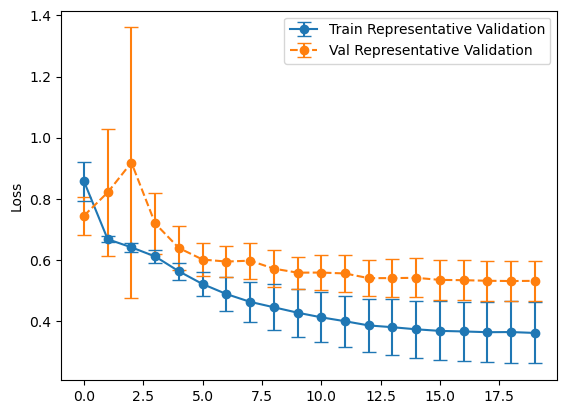
Experiment #1 Evaluation
plot_losses(history_balanced, "Balanced Validation")
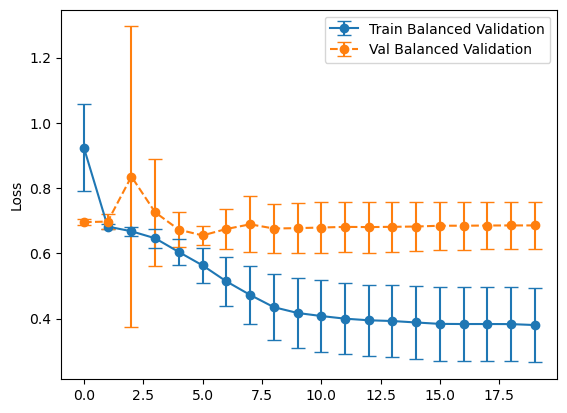
plot_losses(history_representative, "Representative Validation")
balanced_mean_metrics = get_means(evals_balanced)
representative_mean_metrics = get_means(evals_representative)
for name, value in zip(metric_names, balanced_mean_metrics):
print(f"{name}: {value}")
loss: 0.6249180614948273
tp: 58.0
fp: 255.4
tn: 744.6
fn: 42.0
accuracy: 0.7296363592147828
precision: 0.1723228558897972
recall: 0.5799999952316284
for name, value in zip(metric_names, representative_mean_metrics):
print(f"{name}: {value}")
loss: 0.5781383991241456
tp: 67.7
fp: 279.8
tn: 720.2
fn: 32.3
accuracy: 0.7162727266550064
precision: 0.20275740325450897
recall: 0.6770000040531159
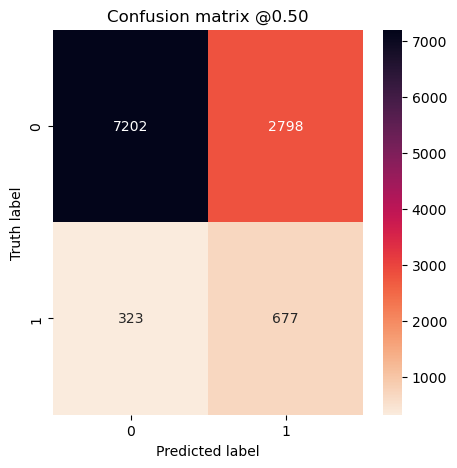
So far it doesn’t seem to matter much. Let’s look at confusion matrices to make sure.
concat_preds_balanced = np.concatenate(preds_balanced)
concat_preds_unbalanced = np.concatenate(preds_representative)
true_labels = tf.concat([y for x, y in test_ds], axis=0)
num_runs = len(preds_balanced)
concat_labels = np.tile(true_labels.numpy(), num_runs)
plot_cm(concat_labels, concat_preds_balanced)
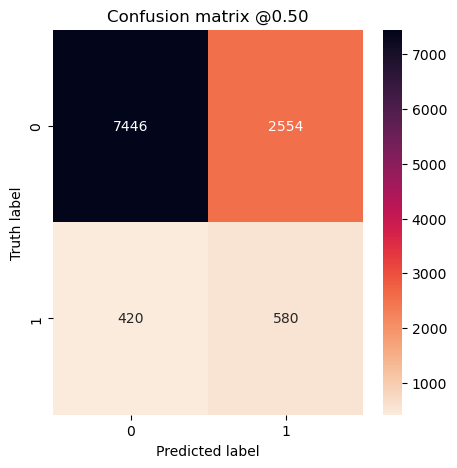
plot_cm(concat_labels, concat_preds_unbalanced)
Experiment #2
In this case we’ll use a higher learning rate. Everything else remains the same.
optimizer = tf.keras.optimizers.Adam(learning_rate=0.003)
history_balanced, preds_balanced, evals_balanced = run_experiment(
train_ds, val_ds_balanced, test_ds, callbacks=callbacks
)
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
history_representative, preds_representative, evals_representative = run_experiment(
train_ds, val_ds_representative, test_ds, callbacks=callbacks
)
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 2s 20ms/step
Experiment #2 Evaluation
plot_losses(history_balanced, "Balanced Validation")
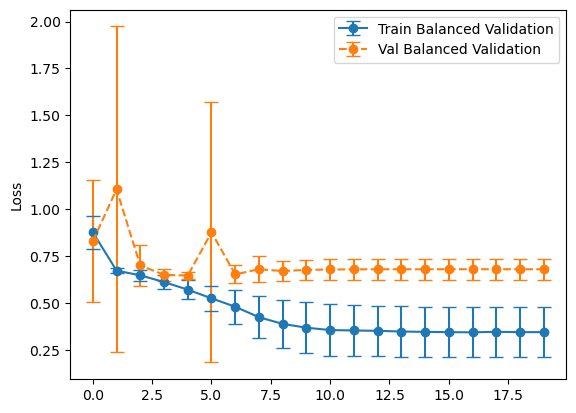
plot_losses(history_representative, "Representative Validation")
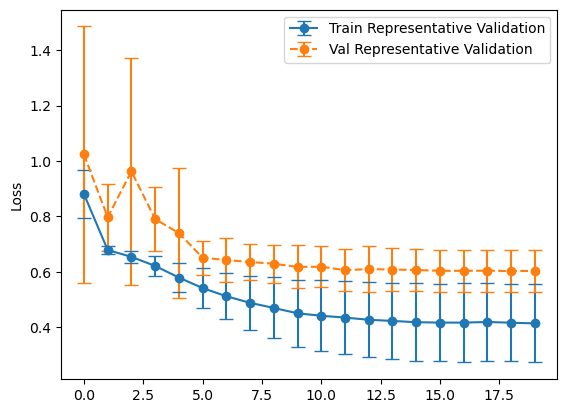
balanced_mean_metrics = get_means(evals_balanced)
representative_mean_metrics = get_means(evals_representative)
for name, value in zip(metric_names, balanced_mean_metrics):
print(f"{name}: {value}")
loss: 0.6253237187862396
tp: 60.6
fp: 234.5
tn: 765.5
fn: 39.4
accuracy: 0.7510000050067902
precision: 0.18702139407396318
recall: 0.6060000061988831
for name, value in zip(metric_names, representative_mean_metrics):
print(f"{name}: {value}")
loss: 0.6437865197658539
tp: 61.9
fp: 285.7
tn: 714.3
fn: 38.1
accuracy: 0.7056363672018051
precision: 0.1674203671514988
recall: 0.6189999938011169
concat_preds_balanced = np.concatenate(preds_balanced)
concat_preds_unbalanced = np.concatenate(preds_representative)
plot_cm(concat_labels, concat_preds_balanced)
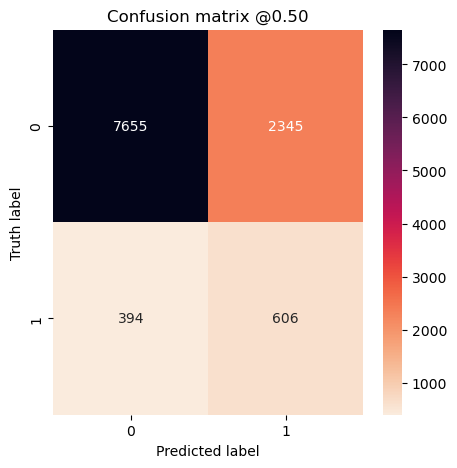
plot_cm(concat_labels, concat_preds_unbalanced)
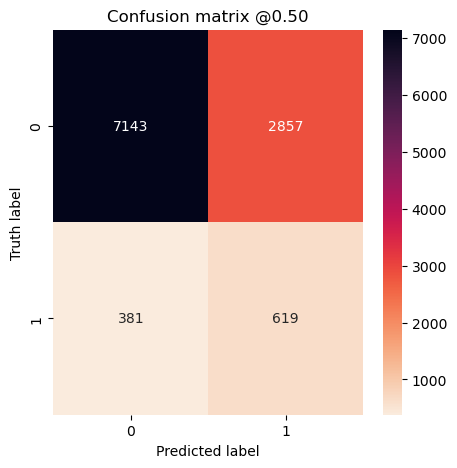
Experiment #3
Once more with a much lower learning rate.
optimizer = tf.keras.optimizers.Adam(learning_rate=0.00008)
history_balanced, preds_balanced, evals_balanced = run_experiment(
train_ds, val_ds_balanced, test_ds, callbacks=callbacks
)
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 2s 22ms/step
history_representative, preds_representative, evals_representative = run_experiment(
train_ds, val_ds_representative, test_ds, callbacks=callbacks
)
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 21ms/step
69/69 [==============================] - 2s 22ms/step
69/69 [==============================] - 2s 20ms/step
69/69 [==============================] - 2s 21ms/step
Experiment #3 Evaluation
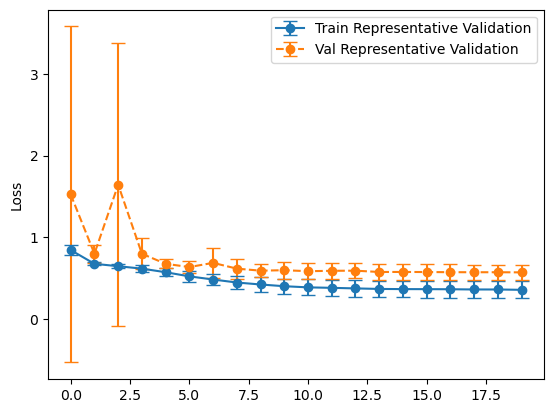
plot_losses(history_balanced, "Balanced Validation")
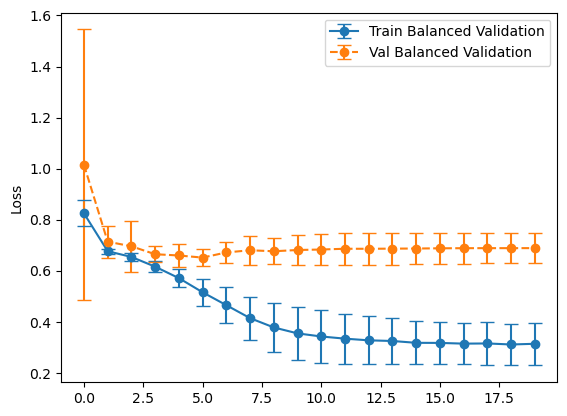
plot_losses(history_representative, "Representative Validation")
balanced_mean_metrics = get_means(evals_balanced)
representative_mean_metrics = get_means(evals_representative)
for name, value in zip(metric_names, balanced_mean_metrics):
print(f"{name}: {value}")
loss: 0.6268290698528289
tp: 67.7
fp: 275.3
tn: 724.7
fn: 32.3
accuracy: 0.7203636288642883
precision: 0.2012877956032753
recall: 0.6769999921321869
for name, value in zip(metric_names, representative_mean_metrics):
print(f"{name}: {value}")
loss: 0.620904928445816
tp: 66.5
fp: 290.9
tn: 709.1
fn: 33.5
accuracy: 0.7050909101963043
precision: 0.1918455943465233
recall: 0.6650000095367432
concat_preds_balanced = np.concatenate(preds_balanced)
concat_preds_unbalanced = np.concatenate(preds_representative)
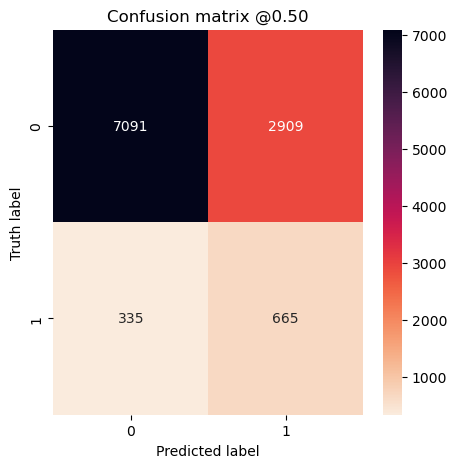
plot_cm(concat_labels, concat_preds_balanced)
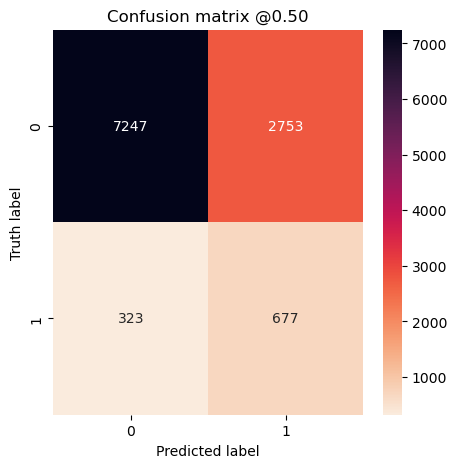
plot_cm(concat_labels, concat_preds_unbalanced)
Round 1 Conclusion
Overall, it looks like the model that used the balanced validation set did better on the test set. This might be a bit of a surprise as validation datasets are supposed representative of the test set, but that one did worse. I think what’s going on is we’re using our validation set for two separate tasks - validating our results and using its loss in the callbacks. I’ve rerun this experiment several times and gotten similar results.
Now let’s try it with a better model. We used a simple model for the first round of experiments. Now let’s do the same thing except using Xception.
Experiment #4
optimizer = tf.keras.optimizers.Adam(learning_rate=0.0003)
history_balanced, preds_balanced, evals_balanced = run_experiment(
train_ds, val_ds_balanced, test_ds, callbacks=callbacks
)
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
history_representative, preds_representative, evals_representative = run_experiment(
train_ds, val_ds_representative, test_ds, callbacks=callbacks
)
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 138ms/step
Experiment #4 Evaluation
plot_losses(history_balanced, "Balanced Validation")
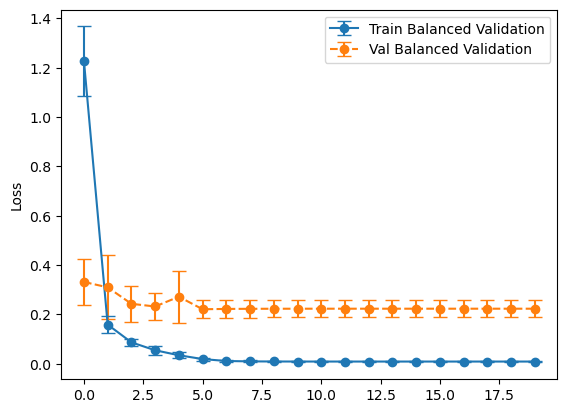
plot_losses(history_representative, "Representative Validation")
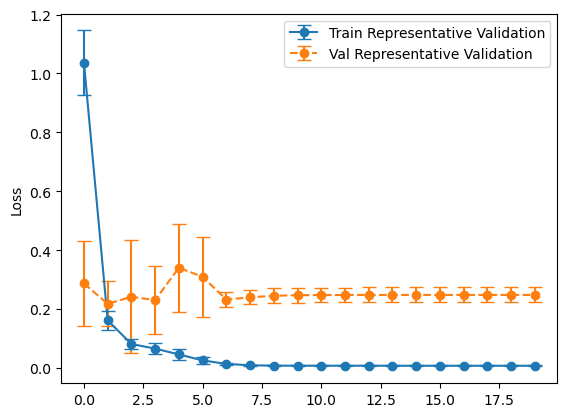
balanced_mean_metrics = get_means(evals_balanced)
representative_mean_metrics = get_means(evals_representative)
for name, value in zip(metric_names, balanced_mean_metrics):
print(f"{name}: {value}")
loss: 0.17637898325920104
tp: 95.3
fp: 48.1
tn: 951.9
fn: 4.7
accuracy: 0.9519999861717224
precision: 0.664779794216156
recall: 0.9529999852180481
for name, value in zip(metric_names, representative_mean_metrics):
print(f"{name}: {value}")
loss: 0.1791197821497917
tp: 95.1
fp: 47.5
tn: 952.5
fn: 4.9
accuracy: 0.9523636221885681
precision: 0.667411994934082
recall: 0.9509999871253967
concat_preds_balanced = np.concatenate(preds_balanced)
concat_preds_unbalanced = np.concatenate(preds_representative)
true_labels = tf.concat([y for x, y in test_ds], axis=0)
num_runs = len(preds_balanced)
concat_labels = np.tile(true_labels.numpy(), num_runs)
plot_cm(concat_labels, concat_preds_balanced)
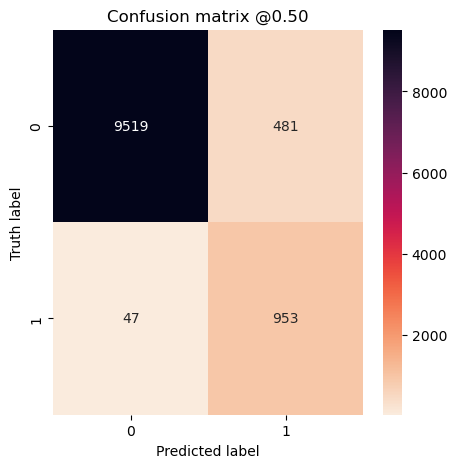
plot_cm(concat_labels, concat_preds_unbalanced)
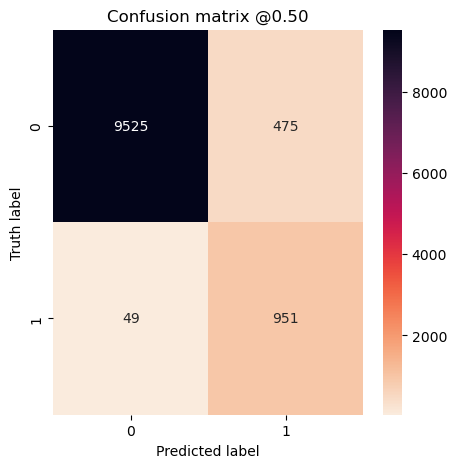
Experiment #5
Using a higher learning rate.
optimizer = tf.keras.optimizers.Adam(learning_rate=0.003)
history_balanced, preds_balanced, evals_balanced = run_experiment(
train_ds, val_ds_balanced, test_ds, callbacks=callbacks
)
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 138ms/step
history_representative, preds_representative, evals_representative = run_experiment(
train_ds, val_ds_representative, test_ds, callbacks=callbacks
)
69/69 [==============================] - 10s 138ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 136ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 136ms/step
69/69 [==============================] - 10s 136ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 137ms/step
69/69 [==============================] - 10s 136ms/step
Experiment #5 Evaluation
plot_losses(history_balanced, "Balanced Validation")
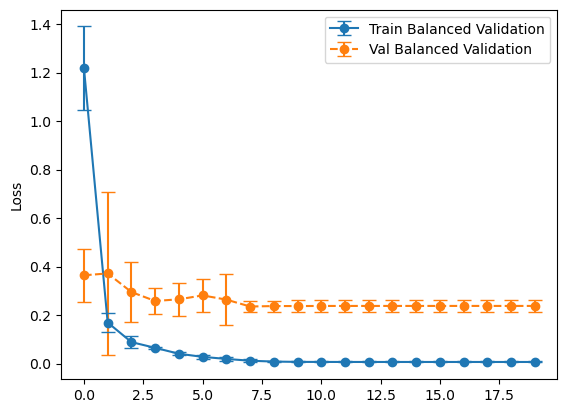
plot_losses(history_representative, "Representative Validation")
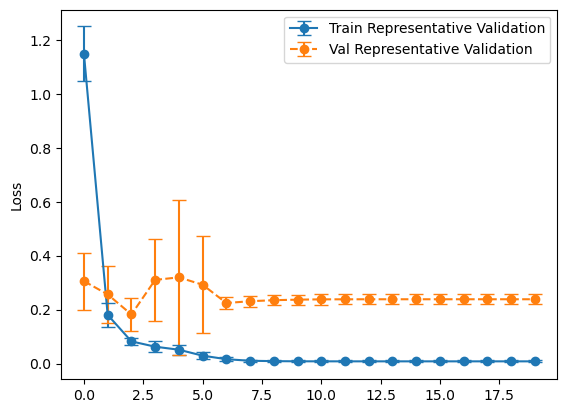
balanced_mean_metrics = get_means(evals_balanced)
representative_mean_metrics = get_means(evals_representative)
for name, value in zip(metric_names, balanced_mean_metrics):
print(f"{name}: {value}")
loss: 0.18235086351633073
tp: 95.1
fp: 46.8
tn: 953.2
fn: 4.9
accuracy: 0.9529999852180481
precision: 0.6707041501998902
recall: 0.9509999871253967
for name, value in zip(metric_names, representative_mean_metrics):
print(f"{name}: {value}")
loss: 0.17296597808599473
tp: 95.0
fp: 47.3
tn: 952.7
fn: 5.0
accuracy: 0.9524545311927796
precision: 0.6679466247558594
recall: 0.949999988079071
concat_preds_balanced = np.concatenate(preds_balanced)
concat_preds_unbalanced = np.concatenate(preds_representative)
plot_cm(concat_labels, concat_preds_balanced)
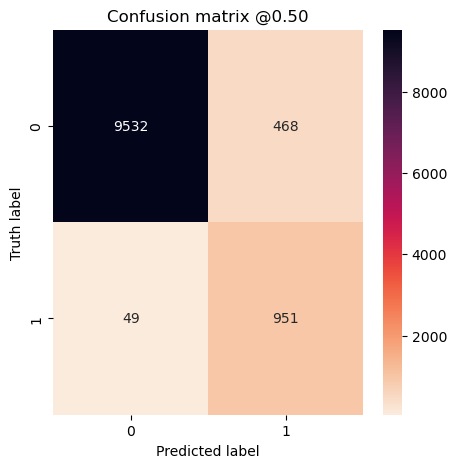
plot_cm(concat_labels, concat_preds_unbalanced)
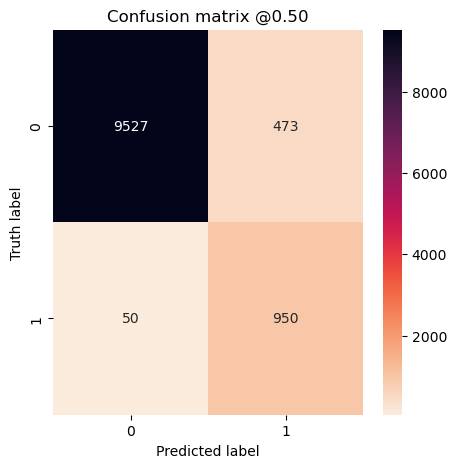
Experiment #6
Once more with a much lower learning rate.
optimizer = tf.keras.optimizers.Adam(learning_rate=0.00008)
history_balanced, preds_balanced, evals_balanced = run_experiment(
train_ds, val_ds_balanced, test_ds, callbacks=callbacks
)
69/69 [==============================] - 10s 136ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 139ms/step
69/69 [==============================] - 10s 141ms/step
history_representative, preds_representative, evals_representative = run_experiment(
train_ds, val_ds_representative, test_ds, callbacks=callbacks
)
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 141ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 13s 138ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 140ms/step
69/69 [==============================] - 10s 141ms/step
Experiment #6 Evaluation
plot_losses(history_balanced, "Balanced Validation")
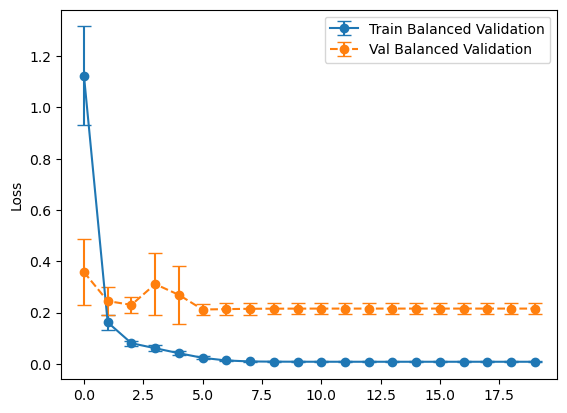
plot_losses(history_representative, "Representative Validation")
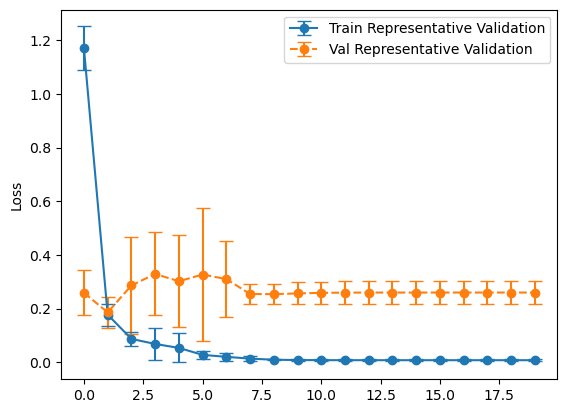
balanced_mean_metrics = get_means(evals_balanced)
representative_mean_metrics = get_means(evals_representative)
for name, value in zip(metric_names, balanced_mean_metrics):
print(f"{name}: {value}")
loss: 0.17486101090908052
tp: 95.2
fp: 46.8
tn: 953.2
fn: 4.8
accuracy: 0.9530908942222596
precision: 0.6706463515758514
recall: 0.9519999861717224
for name, value in zip(metric_names, representative_mean_metrics):
print(f"{name}: {value}")
loss: 0.18693130910396577
tp: 95.7
fp: 48.9
tn: 951.1
fn: 4.3
accuracy: 0.9516363501548767
precision: 0.662167078256607
recall: 0.9569999814033509
concat_preds_balanced = np.concatenate(preds_balanced)
concat_preds_unbalanced = np.concatenate(preds_representative)
plot_cm(concat_labels, concat_preds_balanced)
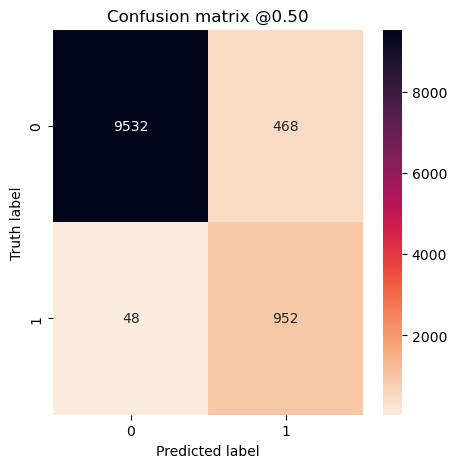
plot_cm(concat_labels, concat_preds_unbalanced)
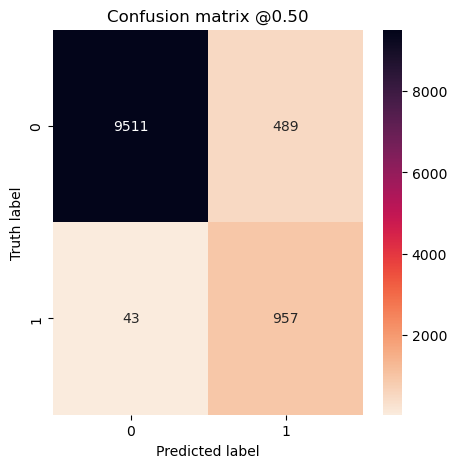
Overall Conclusion
We ran the experiment with Xception three times and each time the models converged to about the same final performance. It seems like with more advanced models, the distribution of the validation set didn’t matter. Because of that, I would make it representative to match the test set.