What goes on inside the black box of a machine learning algorithm? While it may be impossible for a human to understand precisely why a large neural network produced the results it did, some algorithms are far more transparent. Decision trees are just such an example of a machine learning algorithm whose results can be understood by people.
Table of Contents
- Data Exploration
- Data Cleaning
- Preparing the Data
- Building the Model
- Testing the Model
- Visualizing the Model
- Random Forests
- Conclusion
To explore the power of decision trees, I’ll use them to attempt to classify mushrooms into either poisonous or edible based on their look, smell, and habitat. The Audubon Society Field Guide to North American Mushrooms (1981) recorded these attributes for each of 8124 different mushrooms. As well as the toxicity, they came up with 22 other ways to characterize the mushrooms, such as cap shape, odor, and gill size. Credit for publishing the dataset goes to: Dua, D. and Karra Taniskidou, E. (2017). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.
import collections
from collections import defaultdict
import pandas as pd
import pydotplus
from sklearn import tree
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.tree import DecisionTreeClassifier
The attributes are all described on the dataset’s website, but they aren’t included with the dataset. We’ll have to tell pandas to skip the header and we’ll add the column names manually afterward.
df = pd.read_csv('agaricus-lepiota.data', header=None)
Here are the attributes provided. The first is the toxicity, which is what we will be trying to predict from the other attributes.
columns = ['toxicity', 'cap-shape', 'cap-surface', 'cap-color', 'bruises', 'odor', 'gill-attachment', 'gill-spacing', 'gill-size',
'gill-color', 'stalk-shape', 'stalk-root', 'stalk-surface-above-ring', 'stalk-surface-below-ring',
'stalk-color-above-ring', 'stalk-color-below-ring', 'veil-type', 'veil-color', 'ring-number', 'ring-type',
'spore-print-color', 'population', 'habitat']
df.columns = columns
Data Exploration
Now we’ll take a quick look at the data.
df.head()
| toxicity | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | k | s | u |
| 1 | e | x | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | n | g |
| 2 | e | b | s | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 3 | p | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | u |
| 4 | e | x | s | g | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
5 rows × 23 columns
For toxicity, we have ‘p’ for poisonous and ‘e’ for edible. The other attributes are:
- cap-shape: bell=b,conical=c,convex=x,flat=f, knobbed=k,sunken=s
- cap-surface: fibrous=f,grooves=g,scaly=y,smooth=s
- cap-color: brown=n,buff=b,cinnamon=c,gray=g,green=r, pink=p,purple=u,red=e,white=w,yellow=y
- bruises?: bruises=t,no=f
- odor: almond=a,anise=l,creosote=c,fishy=y,foul=f, musty=m,none=n,pungent=p,spicy=s
- gill-attachment: attached=a,descending=d,free=f,notched=n
- gill-spacing: close=c,crowded=w,distant=d
- gill-size: broad=b,narrow=n
- gill-color: black=k,brown=n,buff=b,chocolate=h,gray=g, green=r,orange=o,pink=p,purple=u,red=e, white=w,yellow=y
- stalk-shape: enlarging=e,tapering=t
- stalk-root: bulbous=b,club=c,cup=u,equal=e, rhizomorphs=z,rooted=r,missing=?
- stalk-surface-above-ring: fibrous=f,scaly=y,silky=k,smooth=s
- stalk-surface-below-ring: fibrous=f,scaly=y,silky=k,smooth=s
- stalk-color-above-ring: brown=n,buff=b,cinnamon=c,gray=g,orange=o, pink=p,red=e,white=w,yellow=y
- stalk-color-below-ring: brown=n,buff=b,cinnamon=c,gray=g,orange=o, pink=p,red=e,white=w,yellow=y
- veil-type: partial=p,universal=u
- veil-color: brown=n,orange=o,white=w,yellow=y
- ring-number: none=n,one=o,two=t
- ring-type: cobwebby=c,evanescent=e,flaring=f,large=l, none=n,pendant=p,sheathing=s,zone=z
- spore-print-color: black=k,brown=n,buff=b,chocolate=h,green=r, orange=o,purple=u,white=w,yellow=y
- population: abundant=a,clustered=c,numerous=n, scattered=s,several=v,solitary=y
- habitat: grasses=g,leaves=l,meadows=m,paths=p, urban=u,waste=w,woods=d
df.describe()
| toxicity | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | ... | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 |
| unique | 2 | 6 | 4 | 10 | 2 | 9 | 2 | 2 | 2 | 12 | ... | 4 | 9 | 9 | 1 | 4 | 3 | 5 | 9 | 6 | 7 |
| top | e | x | y | n | f | n | f | c | b | b | ... | s | w | w | p | w | o | p | w | v | d |
| freq | 4208 | 3656 | 3244 | 2284 | 4748 | 3528 | 7914 | 6812 | 5612 | 1728 | ... | 4936 | 4464 | 4384 | 8124 | 7924 | 7488 | 3968 | 2388 | 4040 | 3148 |
4 rows × 23 columns
In total, we have 8124 samples and 22 different attributes (not counting toxicity). That should be enough to train a decision tree.
We also have a good class distribution. The most common toxicity category is ‘e’ for edible, but that’s only 4208 samples. That means that there are 3196 poisonous samples in the dataset. That’s close enough to 50-50, so we’ve got plenty of both categories.
Data Cleaning
The first thing to do is to check for missing data.
df.isnull().sum()
toxicity 0
cap-shape 0
cap-surface 0
cap-color 0
bruises 0
odor 0
gill-attachment 0
gill-spacing 0
gill-size 0
gill-color 0
stalk-shape 0
stalk-root 0
stalk-surface-above-ring 0
stalk-surface-below-ring 0
stalk-color-above-ring 0
stalk-color-below-ring 0
veil-type 0
veil-color 0
ring-number 0
ring-type 0
spore-print-color 0
population 0
habitat 0
dtype: int64
It looks like there are no missing values. Beautiful! That should make things more manageable. We should also go through the data and make sure all the values are what we’re expecting.
for x in range(len(df.columns)):
print(df.columns[x] + ":")
print(df[df.columns[x]].unique())
print("")
toxicity:
['p' 'e']
cap-shape:
['x' 'b' 's' 'f' 'k' 'c']
cap-surface:
['s' 'y' 'f' 'g']
cap-color:
['n' 'y' 'w' 'g' 'e' 'p' 'b' 'u' 'c' 'r']
bruises:
['t' 'f']
odor:
['p' 'a' 'l' 'n' 'f' 'c' 'y' 's' 'm']
gill-attachment:
['f' 'a']
gill-spacing:
['c' 'w']
gill-size:
['n' 'b']
gill-color:
['k' 'n' 'g' 'p' 'w' 'h' 'u' 'e' 'b' 'r' 'y' 'o']
stalk-shape:
['e' 't']
stalk-root:
['e' 'c' 'b' 'r' '?']
stalk-surface-above-ring:
['s' 'f' 'k' 'y']
stalk-surface-below-ring:
['s' 'f' 'y' 'k']
stalk-color-above-ring:
['w' 'g' 'p' 'n' 'b' 'e' 'o' 'c' 'y']
stalk-color-below-ring:
['w' 'p' 'g' 'b' 'n' 'e' 'y' 'o' 'c']
veil-type:
['p']
veil-color:
['w' 'n' 'o' 'y']
ring-number:
['o' 't' 'n']
ring-type:
['p' 'e' 'l' 'f' 'n']
spore-print-color:
['k' 'n' 'u' 'h' 'w' 'r' 'o' 'y' 'b']
population:
['s' 'n' 'a' 'v' 'y' 'c']
habitat:
['u' 'g' 'm' 'd' 'p' 'w' 'l']
They all look good except for stalk root. For some reason, there’s a question mark category. Let’s see how many question marks there are. Hopefully, it’s just one or two, and we can remove those.
len(df[df['stalk-root']=='?'])
2480
OK, it’s a lot. The website says that those values are missing. Because so many samples are affected, I think the best thing to do would be to not include stalk root in the decision tree. Let’s remove it from the dataset.
df = df.drop(['stalk-root'], axis=1)
We’ll also remove the label from the list of columns.
del columns[columns.index('stalk-root')]
Preparing the Data
As we can see, the data are all categorical. We’ll have to convert them to numbers before we use them. Let’s go ahead and do that.
d = defaultdict(LabelEncoder)
# Encoding the variable
numbered_df = df.apply(lambda x: d[x.name].fit_transform(x))
numbered_df.head()
| toxicity | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 5 | 2 | 4 | 1 | 6 | 1 | 0 | 1 | 4 | ... | 2 | 7 | 7 | 0 | 2 | 1 | 4 | 2 | 3 | 5 |
| 1 | 0 | 5 | 2 | 9 | 1 | 0 | 1 | 0 | 0 | 4 | ... | 2 | 7 | 7 | 0 | 2 | 1 | 4 | 3 | 2 | 1 |
| 2 | 0 | 0 | 2 | 8 | 1 | 3 | 1 | 0 | 0 | 5 | ... | 2 | 7 | 7 | 0 | 2 | 1 | 4 | 3 | 2 | 3 |
| 3 | 1 | 5 | 3 | 8 | 1 | 6 | 1 | 0 | 1 | 5 | ... | 2 | 7 | 7 | 0 | 2 | 1 | 4 | 2 | 3 | 5 |
| 4 | 0 | 5 | 2 | 3 | 0 | 5 | 1 | 1 | 0 | 4 | ... | 2 | 7 | 7 | 0 | 2 | 1 | 0 | 3 | 0 | 1 |
5 rows × 22 columns
If we need to, we can always reverse the labeling like this:
# Inverse the encoded
numbered_df.apply(lambda x: d[x.name].inverse_transform(x)).head()
| toxicity | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | k | s | u |
| 1 | e | x | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | n | g |
| 2 | e | b | s | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 3 | p | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | u |
| 4 | e | x | s | g | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
5 rows × 22 columns
Before we feed the data through an algorithm, we’ll have to remove the toxicity because that’s the variables that the algorithms is trying to predict.
labels = numbered_df['toxicity']
samples = numbered_df.drop('toxicity', axis=1)
Now we want to build a model and test it. But to get a fair test of how it would work in the real world, we can’t use any of the testing data for training the model. So we’ll withhold a random 20% of the dataset and train the model with the other 80%.
X_train, X_test, y_train, y_test = train_test_split(samples, labels, test_size = 0.2, random_state = 0)
print("Number of samples to train the model: " + str(len(X_train)))
print("Number of samples to test the model: " + str(len(X_test)))
Number of samples to train the model: 6499
Number of samples to test the model: 1625
Building the Model
Now we’ll build the actual decision tree and train it on the data.
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
DecisionTreeClassifier()
Let’s see how well it did with the data we gave it.
Testing the Model
print("The model is able to categorize {:.0%} of the training set.".format(accuracy_score(y_train, clf.predict(X_train))))
The model is able to categorize 100% of the training set.
Wow. We were able to build a model that accurately categorized every mushroom into either poisonous or edible. That doesn’t mean it can predict 100% of mushroom accurately. To determine how many it can, we’ll have to see how well it does on the mushroom it has never seen before.
print("The model correctly predicted {:.0%} of the test set.".format(accuracy_score(y_test, clf.predict(X_test))))
The model correctly predicted 100% of the test set.
Even more impressive, we were able to correctly predict all 1625 samples that the model hadn’t seen before. That’s a pretty good model. Now, let’s try to visualize it.
Visualizing the Model
We’ll use Graphviz and PyDotPlus to visualize the model.
dot_data = tree.export_graphviz(clf,
feature_names=columns[1:],
out_file=None,
filled=True,
rounded=True,
special_characters=True)
graph = pydotplus.graph_from_dot_data(dot_data)
nodes = graph.get_node_list()
colors = []
for node in nodes:
if node.get_label():
values = [int(ii) for ii in node.get_label().split('value = [')[1].split(']')[0].split(',')]
values = [int(255 * v / sum(values)) for v in values]
color = '#{:02x}{:02x}00'.format(values[1], values[0])
colors.append(color)
node.set_fillcolor(color)
graph.write_png('tree.png')
True
Here is the resulting decision tree. It’s a bit complex, but as it correctly determined the toxicity of every mushroom in the test set, I’m not complaining about it.
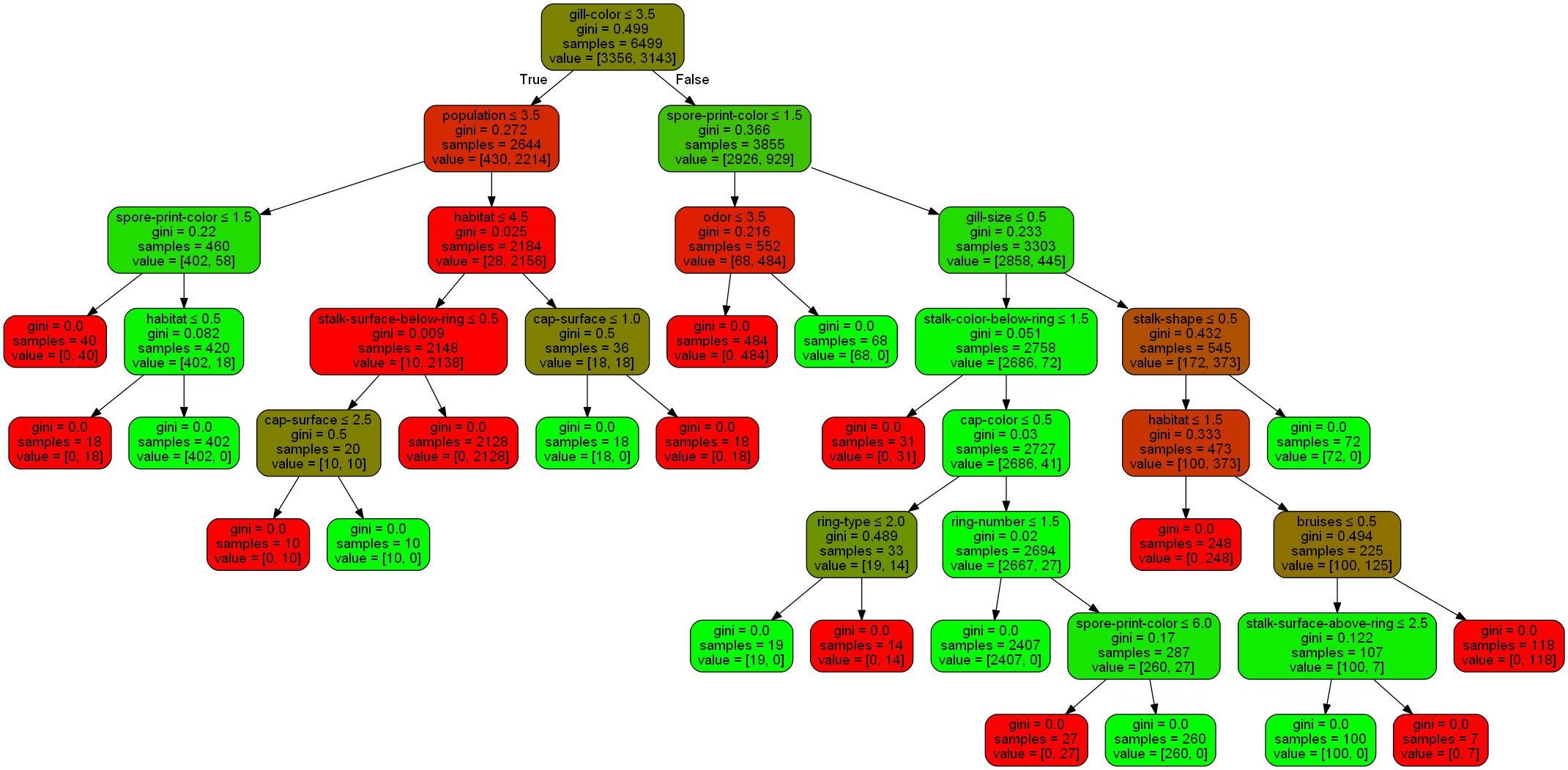
We can learn a lot from this tree. Of the 6499 samples, 3356 of them are edible, and 3143 of them are poisonous. The color of the box corresponds to the distribution of edible versus poisonous. The greener the box, the higher the proportion of those are edible. The redder the box, the higher the proportion are poisonous.
The first thing the decision tree decided to look at was gill color. It first asks the question of whether the gill color is in a category of 3.5 or less. These categories are zero-indexed, so this equates to the first four categories. Here are the categories:
d['gill-color'].classes_
array(['b', 'e', 'g', 'h', 'k', 'n', 'o', 'p', 'r', 'u', 'w', 'y'],
dtype=object)
The first four correspond to buff, red, gray, and chocolate. Out of the 6499 samples that we used to train the decision tree, 2644 had gills of one of these colors, and the other 3855 did not.
Let’s say it isn’t one of those colors, so we go down the “False” path of the tree. Now we look at spore print color and ask if the category is less than 1.5, which would be categories 0 and 1.
d['spore-print-color'].classes_
array(['b', 'h', 'k', 'n', 'o', 'r', 'u', 'w', 'y'], dtype=object)
Those correspond to black and chocolate. Let’s say it is one of those types. So we go down the tree and look at the odor. This is the final deciding factor. Here are the categories for that attribute
d['odor'].classes_
array(['a', 'c', 'f', 'l', 'm', 'n', 'p', 's', 'y'], dtype=object)
From the graph, we can see that if the odor is in a category less than or equal to 3.5, which would be the first four categories, it is poisonous. Those categories are almond, creosote, foul, and anise. If it’s not one of those, the mushroom is edible.
Random Forests
In a more complex scenario where we weren’t able to predict with 100% accuracy from a single tree, we would build a “random forest” by combining many different decision trees. Then the trees can be weighed and combined in various ways to make an overall algorithm. The algorithm becomes harder to visualize at that point, but you can still learn a lot about the data from this technique.
We already know that we can accurately predict every mushroom in the dataset with a single decision tree, but let’s put some constraints on it. Before, we could have as many forks as we wanted, allowing us to build a very complicated decision tree. Now, let’s limit the number of forks to 4 and see how well we can do.
forest_clf = RandomForestClassifier(n_estimators=100, max_depth=4, random_state=0)
forest_clf.fit(X_train, y_train)
print("The random forest model correctly predicted {:.0%} of the test set.".format(accuracy_score(y_test, forest_clf.predict(X_test))))
The random forest model correctly predicted 99% of the test set.
Because of the way random forests bootstrap different subsets of the data, you can’t have an “average” of the random forest that’s a single decision tree. That means there’s no way to visualize a random forest. To get a sense of what the model is doing, we can see what it considers the most important features, and see how what percentage of the predictive power they account for.
feat_importance_dict = dict(zip(columns[1:], forest_clf.feature_importances_))
most_important = sorted(feat_importance_dict, key=feat_importance_dict.get, reverse=True)[:5]
for item in most_important:
print("{item}: {value:.2%}".format(item=item, value=feat_importance_dict[item]))
odor: 17.14%
gill-size: 15.81%
gill-color: 14.05%
spore-print-color: 11.51%
ring-type: 6.67%
So it turns out that odor is most important, followed by gill size and gill color.
Conclusion
Decision trees are incredible algorithms and have the wonderful attribute that they can be understood by people. Even more, they can be combined together to form powerful random forest classifiers. Random forests are often considered one of the most important machine learning algorithms and are also one of the most powerful. In fact, a study in 2014 tested 179 algorithms and determined that the random forest was the best overall algorithm.